AAs you know, different markers (or SNP mutations) are used to identify the haplogroups into which we classify human mtDNA these markers are assumed to have arisen by chance in a purely random manner within a population, and that they are not lost as they are passed on from mother to daughter generation after generation. This allows us to trace the relationship between different populations by verifying which markers are present and which are not.
Markers are therefore mutations, and they can be found in different regions of the human mtDNA. Today's post will look at the mtDNA's "Coding Region" and in a very particular mutation which is actually a deletion (part of the sequence found in some populations is absent in those carrying the deletion), which was lost due to the random mutation: the 9-bp deletion (9-bp d).
The 9-bp d in different populations around the world
First we will define the 9-bp deletion as a "length variant in the intergenic region between cytocrome c oxidase subunit II and the mitochondrial tRNA for lysine (COII⁄tRNALys)" [3]. What this means in a physically sense is the loss (deletion) of one (1) of the two (2) tandem repeats of the sequence "CCCCCTCTA" between nps 8281 and 8289. It is known also as the polymorphic anthropological marker MIC9D.
On the 9-bp sequence
Most people carry two (2) copies of the 9-bp sequence "CCCCCTCTA". This is generally assumed by mainstream science to be the ancestral state.
State which can be modified by mutations such as insertions of one or two additional 9-bp sequences, or by repeats of additional cytosine (C) or deletions and also by the loss of one full sequence (the 9-bp deletion). [12]
It was first detected among Asians by Wrischnik et al. (1987) [4], but later it was found at high frequencies (77% to 100%) in other populations in Polynesia such as the Maori, Niuean, Samoan and Hawaiian as well as among Australian Aboriginals:
The Polynesian branch seems to be associated with the influx or people from Taiwan who migrated southwards across Indonesia and led to the peopling of Polynesia. [5]
Another region where it is found is in the Indian subcontinent and insular areas next to it, where it is carried by southern Indian populations at frequencies that range from 68% in Yandi and 44% in Irula to 24% in Nicobarese and 4% in Siddi, [3] and 21% in Koyas and 3% in Chenchus. [2]
The Asian variant is found at frequencies of 15.6% (Thailand), 40% (Taiwanese Han), 20% (Hong Kong and Shanghai), but 3.3% in Shandong and Uygur populations. [5] In China it averages 14.7% (ranging from 0% to 32% in different ethnic groups). [15]
In addition to these Asian groups, it is also found in Africa and the Americas (as I said, it is global)
Africa
Among sub-Saharan Africans it is found at a frequency of 8.8% (or 81 individuals out of n=919), who carried it in different haplotypes (41 haplotypes in total).
The African Khoisan do not carry it, and is not frequent or absent among the people of the Western and Southwestern Africa. Pygmies and other populations in Central Africa, Malawi and Southern Bantu-speakers do carry it, suggesting according to orthodoxy a Central African origin and a recent dispersal to other regions. [1]
America
In America it is found in those populations belonging to mitochondrial haplogroup B, where it serves as a marker. It is one of the four founding lineages of Native Americans, but also has the peculiarity of being the only mtDNA haplogroup to cover the Pacific: it is found on both shores of the Pacific Ocean, in North and South America, Polynesia, Indonesia, Thailand, the Philippines, China and Japan.
Global yet different at a regional level
This global range is quite unusual since it spans different haplogroups.
The African and Asian (East Asia and India) mtDNA haplotypes are distinct and separate (see Fig. 2 in [3]).
A later study by Kivisild et al., (2003) [2] covering certain Indian populations noted that their 9-bp d was different in its haplotypes to the other Asian and African deleted lineages.
To study the relationship between the different populations carrying the 9-bp d, the following tree (Fig. 4 in [5]) is very useful. It shows that Africans (both those with the deleted 9-bp and the non-deleted populations) cluster together away from the rest.
The Australian Aboriginals and Indians without the 9-bp d are also distinctly separated on the tree from Indians and Australians carrying it.
The Chinese "cluster I" is very close to the non-deleted Chinese, which suggests a recent local origin.
The Amerindians and deleted Amerindians are very far apart.
There is a distinct separation which has three branches: (a) with all of the 9-bp deleted Thai, Cambodian and Nicobarese, (2) the Taiwan aboriginal, Amerindians, PNG, Indonesians and Polynesians and (c) the Chinese "cluster II".
The tree which does not specify the Haplogroups of the different populations clearly shows thre clusters which display the proximity of Amerindians, Asians and Melanesians; of Africans and of Aboriginal Australians.
Diseases linked to the 9-bp locus deletion
The locus of this 9-bp segment seems to have some strong correlation with the individuals' health: A study by Yiqui Jin et al, (2012) [6], links it to a form of cancer (Hepatocellular carcinoma or HCC). This is a very common form of cancer worldwide. The scholars found that the presence of the 9-bp deletion increases the risk of cancer (they call the deletion "one repeat", which is technically correct since the normal form is a tandem repeat [two repeats], so a deletion menas a "one repeat"):
"carriage of 9-bp one repeat fragment was associated with a significantly increased risk of developing HCC
[...] the frequency of 9-bp one repeat [that is: deletion] in our control samples (16.0%) was similar to that of previous reports in Chinese (14.7%). However, this frequency was 22.1% in the HCC groups, indicating a significant association between the 9-bp one repeat and the incidence of HCC." [6]
The reason for this is somehow linked to microRNA (miR) -mediated regulation because the deletion lies in a binding site region for certain miRs.
The locus is also associated with several other diseases: ataxias, idiopathic disorders, polycystic ovary syndrome, dilated cardiomyopathy, etc. [7].
If there is disease risk associated to the presence of one repeat, it seems logical to assume that natural selection would act to eliminate this mutation whenever it appears. However this does not appear to be the case, it is found in old (> 50 ky old haplogroups) with a global range.
Why? The deletion must provide its carriers with a positive side effect. But I have not managed to find any paper mentioning it.
The deletion and its origin
We have seen above that it appears all across the World (though at very low frequencies in Europe) and is considered by orthodox science as a marker that appeared independently in different human populations, yet despite this mutability it is neverheless used as a marker to define mtDNA haplotypes!:
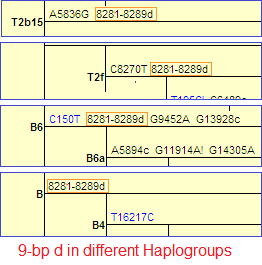
It appears in several haplotypes as markers in some, as a mutation in others:
- B [9]
- B6 [9]
- T2b15 [9]
- T2f [9]
- L0a2 [10]
- W [11]
- H [15]
the fact that it has such a high recurrence, meaning that it is found in different populations and in different mtDNA haplogroups around the world is very odd for a marker, since we would expect it to be distinct and appear only once and remain immutable.
Furthermore, the 9-bp d appears in the Coding Region and "Coding-region SNPs, [are] used as reliable markers to define Hgs because they are considered stable evolutionary events" [8]. But as I pointed out above, it seems that they "are nevertheless not entirely stable" [8] and a paper (Behar et al., 2007 [8]) shows that this instability leads to "inconsistent" haplogroup typing in 3.2% of the samples:
The 9-bp deletion is the major contributor to these inconsistencies, with an overall frequency of 1.2%. Which is quite logical since as we have seen above it appears in at least 7 haplotypes, and if the sequencing does not cover many markers an incorrect haplo-typing is surely to occur. [8]
Origin
According to Sodyall, Vigilant, Hill, Stonekin and Jenkins, 1996, [1] it arose independently in different populations around the world: the "locations and frequencies of variant sites" in Africans and Asians are not the same (different haplotypes), which suggests that ir appeared independently in both regions, and perhaps more than once in Africa [1] and twice in Asia [12].
Behar et al., 2007, also interpret this 9-bp deletion as a Homoplasy (which combines the Greek words for "same" -homo- and "form, shape" -plasis), in other words the chance duplication of a feature with a similar shape, structure or function but with an independent origin and no shared ancestry.
Homoplasy is found in the shape of the fins of fish, dolphins and Ichthyosaurs or wings in bats, birds, pterodactyls; which arose independently in separate and separate lineages of different kinds of creatures (reptiles, fish, mammals and birds) by chance, yet the shape is similar because of their similar function.
The independent origin of the 9-bp d is supported by the fact that the "most likely mode of origin of the deletion, namely, slipped mispairing during replication, can occur (and recur) in every population..." [12].
However Kivisild et al., (2003) [2], apart from suggesting an independent origin of the deletion also consider the possibility of "an ancient common root" [2]. And this is indeed interesting, so we will look into it below.
Ancient relict state
Behar et al., [8] found that out of a total amount of mutations of 592, there were 343 transitions, 199 tranversions, 35 insertions and 15 deletions. So the most rare mutation is a deletion (2,5% frequency). Yet for mainstream science, the deletion is the most obvious cause for the presence of one instead of two "CCCCCTCTA" sequences. But insertions are over twice as frequent as deletions. And in fact the tandem repeat is more common than the one-repeat (or deletion). Furhtermore, as we will see below, triplicate, cuadruplicate and even intermediate (expansion and contraction of the tandem repeat by loss or addition of individual bases) are present. What does not exist is a total deletion of the segment implying that the trend is towards growth not contraction.
Its low frequencies may be due to its association with illness, but since it has not been selected to extinction, it must provide some benefit to those carrying it.
The fact that it is found repeatedly in several haplotpes and especially in the ancestral African L0a2 line is an indication of its ancient origin. The most parsimonious explanation is that it appeared in the African populations (in fact their placement on the tree above, away from most groups clearly shows a distinct origin) and was lost in some of the downstream haplogroups due to a "9-bp insertion" that erased the "one repeat" and created the now most common tandem repeat, found in most populations. It is likely that a small number of individuals conserved the archaic deleted state. Which, as we see above makes its carrier prone to certain diseases and therefore maintained a low frequency of this variant.
It moved with humans into Asia, generating the Indian - Australian cluster and then (this resembles the Y chromosome C hg. expansion!) into South East Asia, Eastern Asia, Siberia and America.
It grew among mtDNA haplogroup B in such a way as to become its marker. This must imply some yet unknown benefit in carrying the 9-bp deletion.
Mutations
The Sakha from Siberia have a high frequency of mutation associated to the 9-bp locus: 1.8% deletions, 1.2% triplications and 0.26% four repeats all others have the usual "tandem repeat". These mutations were found among different mtDNA haplogroups, where B, T and W were the most common for deletions. [11]
Interestingly "No neighboring populations have been reported to carry a non-haplogroup B deletion [...] suggesting that shared ancestry or admixture or both are unlikely explanations for the presence of these polymorphisms in the Sakha. The identification of high levels of variation may be a function of the large sample size [N = 779] and the in-depth analysis of all derived polymorphisms" [11].
The comment on sample size is a very positive one, in my opinion most studies are based on very small samples and their sequencing is not done thoroughly, so many differences go unnoticed.
The authors conclude that these mutations are due to "multiple recent events" [11], but in my opinion the lack of admixture or shared ancestry means that these are archaic or relict mutations that have been carried by the Sakha and their ancestors for many generations.
The "three repeat" is very infrequent globally, besides the Sakha with 1.2% frequency, the triplicate insertion has been found among Chuckchi (Siberia), Nepalese Tharu (in 1 out of N=127) [12][13], a Portuguese Brazilian belonging to hg. H (1 in N=245), and Portuguese in Europe (2 in N=96) [14]; it is also very rare among Chinese at 0.25% (3 in N=1218) [15].
The normal tandem (two repeat) is also found mutated in some Indian populations. It appears as "contracted" among the Caste populations (Brahmin and Jalari) at an average frequency of 1% and as an "expanded" two repeat mutation among tribal populations (Khonda Dora and Santal) with a 3% frequency. [3] The expansion is due to a T to C transition followed by a replication of the C component. The contractions have "length variant intermediate between 1 - 2 repeats in length". [3] They have also been detected in Borneo, the Philippines and Samoa. [3]
Selective pressure is surely acting on this ancient mutation maintaining it despite the current prevalence of the tandem sequence.
Closing comments
This is a trait found in the most ancient lineages in Africa and also among other populations at low frequency. Yet one of Amerindians founding mtDNA haplogroups is identified with this specific mutation and it is found at very high frequencies among native Americans.
It causes some propensity to certain diseases yet despite this fact it has not disappeared, this implies that it has some yet to be detected positive aspect for those carrying it.
Deletions are infrequent yet this one is global. It surely signals its ancient origin. Furthermore, its presence in seven different haplotypes indicates in my opinion that the basic axioms of the mtDNA clock and neutral mutations used to set it are not to be taken seriously.
Its dispersion across the globe mimics that of Y chromosome hg. C, which I have suggested may be an ancient (Homo erectus) lineage and much older than mainstream science suggests. Could the 9-bp deletion be an archaic hominin trait?
Clearly some more research is needed here. I will keep my eyes open for additional data.
Sources
[1] H Soodyall, L Vigilant, AV Hill, M Stoneking, T Jenkins, (1996). mtDNA control-region sequence variation suggests multiple independent origins of an “Asian-specific” 9-bp deletion in sub-Saharan Africans. Am J Hum Genet, 58 (1996), pp. 595–608
[2] Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, et al., (2003). The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet 72: 313–332. doi: 10.1086/346068
[3] Watkins W. S. et al., (1999). Multiple origins for the mtDNA 9-bp Deletion in Populations of South India. American Journal of Physical Anthropology 109:147-158
[4] Wrischnik LA, Higuchi RG, Stoneking M, Erlich HA, Arnheim N, Wilson AC., (1987). Length mutations in human mitochondrial DNA; direct sequencing of enzymatically amplified DNA. Nucleic Acids Res 15:529–542
[5] Yong-Gang Yao, W.S. Watkins and Ya-Ping Zhang, (2000). Evolutionary History of the mtDNA 9-bp deletion in Chinese populations and its relevance to the peopling of east and southeast Asia. Hum Geet (2000) 107:504-512 doi: 10.1007/s004390000403
[6] Jin Y., et al., (2012). The mitochondrial DNA 9-bp deletion polymorphism is a risk factor for hepatocellular carcinoma in the Chinese population. Genet Test Mol Biomarkers. 2012 May;16(5):330-4. doi: 10.1089/gtmb.2011.0208. Epub 2012 Jan 27
[7] Borgione E., et al., (2013). The 9-bp deletion in region V of mtDNA: a risk factor of hearing loss and encephalomyopathy in Caucasian populations?. Neurol Sci (2013) 3$:1223-1226 doi: 10.1007/s10072-013-1297-9.
[8] Behar DM, Rosset S, Blue-Smith J, Balanovsky O, Tzur S, et al., (2007). The Genographic Project Public Participation Mitochondrial DNA Database PLoS Genet 3(6): e104. doi:10.1371/journal.pgen.0030104
[9] www.phylotree.org
[10] www.dnahaplogroups.org
[11] L. Tarskaia, R. R. Gray, B. Burkley and C. J. Mulligan, (2006), Genetic variation at the mitochondrial DNA 9-bp repeat locus in the Sakha of Siberia. Human Biology 05/2006; 78(2):179-98. doi: 10.1353/hub.2006.0038
[12] Redd A. J., et al., (1995). Evolutionary History of the COII⁄tRNALys Intergenic 9 base pair deletion in human mitochondrial DNAs from the Pacific. Mol. Biol. Evol. 12(4):604-615.
[13] Passarino G1, Semino O, Modiano G, Santachiara-Benerecetti AS, (1993). COII/tRNA(Lys) intergenic 9-bp deletion and other mtDNA markers clearly reveal that the Tharus (southern Nepal) have Oriental affinities. Am J Hum Genet. 1993 Sep;53(3):609-18.
[14] Alves-Silva J., Guimaraes P., Rocha J., Pena S. and Prado V., (1998). Identification in Portugal and Brazil of a mtDNA Lineage containing a 9-bp triplication of the intergenic COII⁄tRNALys Region Hum Hered 1999,49:56-58
[15] Yao YG1, Watkins WS, Zhang YP, (2000). Evolutionary history of the mtDNA 9-bp deletion in Chinese populations and its relevance to the peopling of east and southeast Asia. Hum Genet. 2000 Nov;107(5):504-12.
Patagonian Monsters - Cryptozoology, Myths & legends in Patagonia Copyright 2009-2014 by Austin Whittall ©

















No comments:
Post a Comment