A paper (Hai et al., 2012) [1] analized a very special and unconventional gene, miR-941, it immediately drew my attention because it goes against the usual decreasing gradient of variability from Africa outwards, which shows lower variability in the fringe regions that are further away from our purported Eden in Africa. This post will look into this oddity and its implications.
microRNAs or miRNA
Genes usually are in charge of producing proteins or RNA strands (i.e. tRNA) that in turn interact with gene expression pathways. But this is not the case with micro RNAs (miRNAs) which have very important roles in gene expression. miRNAs are short single-strands of RNA (20 to 24 nucleotides long), hence the name "micro".
The appearance of a new miRNA can impact on the expression of hundreds of genes, so for this reason, the the authors of the paper [1] set out to detect a human-specific miRNA and then tried to figure out its evolution and impact on modern humans. They sought microRNAs (miRNAs) specific to the human genome.
The older (archaic or more ancient) miRNAs tend to have higher expression levels than those that have appeared more recently, and a survey of several human-specific miRNA showed just that: low expression levels, a sure signal that they are quite recent. But there was an exception: miR-941 which was found in the human brain at very high expression levels, higher than other miRNAs found in humans and primates.
miR-941
More interesting was the fact that its expression was absent in macaques and chimpanzees showing that it evolved in the hominid line after we split from the chimps some 6 million years ago (Mya). Its upper limit is given by Denisovans, who do also carried it, and this split took place about 1 Mya. So the miR-941 evolved in our hominin ancestrors between 6 and 1 Mya.
What is exciting is the fact that it interacts with our brains, and the human intellect began to evolve during that time span: the "Human-specific effects of miR-941 regulation are detectable in the brain and affect genes involved in neurotransmitter signalling." [1] it is active in the cerebellum and the prefrontal cortex, which are two processing regions within our brains.
The media hyped up this paper based on its role as "making us human" and its impact on our brain-power, but, as we will see below its role is much deeper than that.
The paper suggests that our extended life span and a higher tendency to developing cancer (compared to chimpanzees and macaques) is probably due to this microRNA. So we should thank miR-941 not only our higher reasoning abilities but also our unusually long life span (yes, cancer is a terrible collateral effect). The authors put it this way:
"It is, therefore, appealing to speculate that emergence of miR-941 enhanced the maintenance of adult stem cell populations, thus supporting longer human lifespan, but rendering human cells more prone to malignant transformation. The role of miR-941 in the regulation of insulin signaling adds support to this notion. The insulin-signaling pathway was consistently implicated in lifespan regulation in many species, including humans. Notably, experimentally verified targets of miR-941 within this pathway include genes directly shown to be involved in lifespan extension in model organisms: IRS1, PPARGC1A and FOXO140 (ref. 40). Furthermore, FOXO1 was linked to extended human longevity." [1]
Not the usual cline though
Unlike many genetic markers which show a high diversity in Africa and then a decreasing gradient as you move away from our purported homeland, miR-941 shows quite a different picture:
Although it is found in all humans, the number of precursor copy-number, or repeats is quite variable: it ranges from 2 to 11 copies. This variaton is linked to geographical location too. Although there is a clear indication that the usual decreasing cline is not present in miR-941, orthodoxy compels the authors to affirm that there is a higher African variability and a West to East cline!:
"The average pre-miR-941 copy number decreased from the west to the east: from eight copies in sub-Saharan Africans to six copies in Eastern Asians. miR-941 precursor copy number variation was also significantly higher in sub-Saharan Africans compared with 'out of Africa' populations, with the exception of Oceanians and native Americans
[modern human carry] 2–11 copies of miR-941 precursor, with an average of 8 copies found in sub-Saharian Africa, an average of 7 copies in Europe, America, Oceania and most of Asia, and an average of 6 copies in East Asia." [1]
Look at the figure below (from [1]) and make up your own mind; it shows miR-941 precursor copy repeats in different populations:
The upper part of the image (c) shows that the variability is highest among South American Karitiana (5 to 9 counts), Papuans and Palestinians (4 to 7) folllowed by San (6 to 9). In other words, it is not higest among Sub-Saharan Aficans!
In the case of the Amerindians, it is a surprising find since they are, according to orthodoxy, supposedly a bottleneck population "lacking the diversity of the Africans" , the other Eurasian people -purported ancestors of the American Natives have less variability than Amerindians.
Part d reflects this, showing the average of each region with the highest count numbers in Oceania, Africa and among Native Americans. All other populations have a lower count.
The Variance (in part e) clearly shows the highest variances in Africans and Native Americans. This does not depict a west - east cline, actually it drops off in the middle and grows again as you move on east.
This is clearly at odds with the accepted notion that diversity drops off with distance to Africa, with the New World populations being the less diverse! The authors actually state this mainstream notion in their abstract: "... shows a trend for decreasing copy-number with migration out of Africa" [1] which is not what the facts in their paper shows. (preconceptions are so hard to erase aren't they?).
So even though the main text shows that diversity drops in Eurasia but increases in Oceania and America the authors overlook it and don't even try to seek an explanation for it.
Speculations and Copy-number variation (CNV)
The paper points avoids controversy and does not analyse the significanse of the different Copy-numbers, it merely points out that a different amount of repeats "is not unexpected, given general instability of genomic regions formed by tandem repeats" [1].
But it is not chance that is at play here but natural selection the authors point out tha when a new miRNA appears (obviously its initial appearance is due to chance), it may wreak havoc in the established network due to negative changes in gene expression. So the forces in natural selection would quickly get rid of it or modify the binding sites that produce negative effects. In the case of miR-941, since it was not eliminated, what changed were its binding sites, which were lost.
So if Amerindians have a higher Copy-number variation (CNV), there must be a selective pressure at work. Actually this is the case:
"The "high-degree of copy-number variation (CNV) ... among contemporary human ethnic populations, suggest[s] a high degree of functional variability even among humans. New cell biological funcions acquired because of the appearance of mR-941, for example, new controls over sonic hedgehog and insulin-signaling pathways (Hu et al., 2012), are likely to be important as CNV-dependent determinants of human-specific adaptations to the environment and to disease." [2]
In other words, CNV is important and should not be overlooked.
It is clear that Amerindians and Papuans have a wide range of CNV than Europeans or Asians, and comparable to those of Africans. Yet they are always depicted a population that originated from a founder effect and later went through a bottle neck which reduced its genetic diversity even further. Why is CNV higher among them?
I am at a loss to explain this, but it the high CNV in Oceanians and American natives is a well known phenomenon, and is an incongruity that is explained away as a sampling bias! [3]:
"higher-frequency CNVs were more common especially in Oceania and the Americas... in contrast with their usual reduced variation, populations from Oceania and the Americas had more CNV loci and more previously unobserved loci than most other populations." [3]
The authors try to explain this anomaly as due to "some bias may exist in CNV detection" and go into an elaborate explanation on sampling. Then they simply ignore this large variability in CNVs compared to the usual Out of Africa cline of decreasing diversity and conclude that: [3]
"despite a difference from SNPs in the frequency spectrum of the copy-number-variants (CNVs) detected - including a comparatively large number of CNVs in previously unexamined populations from Oceania and the Americas- the global distribution of CNVs largely accords with population analyses for SNP data sets of similar size..." [3]
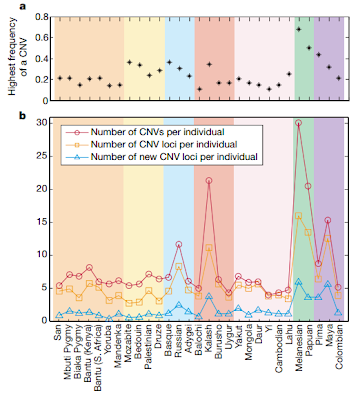
The image below, from [3] actually shows the opposite (peaks in America, Oceania and the Kalash -from Pakistan):
Since African CNV is low, it shows that the high CNV in the Americas is not due to admixture with Africans brought to the New World as slaves. It is due to another cause.
So why do Papuans and Amerindians have a higer CNV diversity?
CNVs can have a negative effect on health and provoke diseases (these will tend to be selected against as the carriers will have a burden which may impede them from having children). Other CNVs have a positive effect and may protect the carrier against disease and "other copy number variants carried by healthy individuals that seem to have no function might actually be evolutionarily retained in populations if they provide a selective advantage." [5]
Could the American and Oceanian variety in CNVs be due to the forces of natural selection retaining CNVs that have been lost by other populations?
Or did they receive these variants from other archaic populations that lived in these areas and admixed with them, receiving these extra CNVs which conferred them a selective advantage?
If so, then the archaic people were already living in America and Sahul when humans reached those regions.
More on the decreasing diverstity myth
When a population with a given diversity (Population A in the image above; diversity is shown by the different colored circles) splits and a sub-population (Population B) moves away, -shown in stage 1- diversity will decrease. In this case we took B as having the same mix as A, but it could have been different, reducing diversity even further (see Population C for instance). The smaller population is more prone to genetic drift which reduces diversity and will also be less likely to evolve new mutations as its size is smaller. It will also be subject to the risk of extinction or loss due to natural calamities than a larger population spread out over a wider territory. So even if the migrating group grows at the same pace as the original population (stages 2 and 3), the outcome will be a reduced diversity.
The acepted theory suggests that variation within a population is proportional only to n (the effective population size) and μ (the mutation rate) which should be the same across all human groups (is it?). [4] So size influences variation: bigger sizes, more variation. Which makes me ask (as an explanation to the higher CNV observed in the New World and Sahul): Did America and Oceania have a larger population in the past which later became extinct? Well, we know that the Americas suffered a devastating impact after its discovery by Europeans in 1492: its population was decimated and its diversity plummeted. Did something simila happen in Oceania? Probably not, as an Old World population they were well adapted to Old World diseases.
Another factor is that, before mutation-drift equilibrium is reached, the age of a population influences its diversity: older populations accumulate more variations. So could the observed CNV diversity be due to an early peopling of America or Oceania by one of our ancestors (Homo erectus? Neanderthal? Denisovans?). This would extend the timeline well beyond the accepted 50 kya for the OoA event or the 15 kya for the peopling of America and allow for diversity to develop.
And, as a closing grand finale look at these two images from Seielstad et al., (1999) [4], which also depict a close relationship between Africans and Americans instead of the usual "decreasing gradient away from Africa":
Yes, the paper (which looks into Y chromosome STR diversity), dates back to 1999 and its figures are extremely unsual but the paper nevertheless echoes the well known mantra of orthodoxy:
"A recent African origin for all humans is supported by a robust corpus of evidence. This evidence takes three major tracks: (1) increased genetic diversity in Africa versus the rest of the world, (2) phylogenetic analyses placing the deepest branches between African and non-African populations, and (3) indications that the age of the “genetic most recent common ancestor” is very young. The Y chromosome, like mtDNA and the autosomes, appears to support a recent African origin." [4]
Sources
[1] Hai Yang Hu, et al., (2012). Evolution of the human-specific microRNA miR-941. Nature Communications 3, Article number: 1145 doi:10.1038/ncomms2146
[2] Antonio Noronha, Changhai Cui, Robert Adron Harris, John C. Crabbe, editors. (2014). Neurobiology of Alcohol Dependence. Elsevier, May 2, 2014. pp 500
[3] Mattias Jakobsson et al., (2008) Genotype, haplotype and copy-number variation in worldwide human populations. vol. 451 doi:10.1038/nature06742
[4] Mark Seielstad, Endashaw Bekele, Muntaser Ibrahim, Amadou Touré, and Mamadou Traoré, (1999). A View of Modern Human Origins from Y Chromosome Microsatellite Variation. Genome Res. Jun 1999; 9(6): 558–567.
[5] Lobo, I. (2008) Copy number variation and genetic disease. Nature Education 1(1):65
Patagonian Monsters - Cryptozoology, Myths & legends in Patagonia Copyright 2009-2014 by Austin Whittall ©



















No comments:
Post a Comment