Y chromosome Haplogroup C is perhaps the oldest and therefore earliest indicator of the presence of modern human males in Eastern Asia. It is found at a relatively low frequencies across most of Asia, reaching its highest frequencies in North Eastern Asia (Mongolia, Siberia); it declines in Japan, Korea, China, India, Southern Asia, but risies in Eastern Indonesia, Polynesia and Australia. It is absent in Africa, found at moderate frequencies in North America and very low frequencies in Europe and South America. [1] (see map below)
As we will see below, it has distinct geographically-specific haplotypes of which C3 is the most widespread one spanning Asia and America.
This extended range and low frequency coverage underlying more recent Eurasian haplogroups clearly indicate that it arose very early in Asia, among first humans that left Africa, during their trek across Western Asia and before they reached Eastern Asia.
In today's post I will summarize its regional haplotypes and frequencies and, close the post with a discussion on their origin and dispersal. I will go over the data and the "orthodox" point of view and later will suggest some non-orthodox ideas regarding C hg. and its origin. To skip the region by region information click here to go to the analysis part of this post.
A Regional Analysis of NRY Haplogroup C
We will detail the C haplogroup's haplotypes and their frequencies of in the following regions:
Australia, Melanesia, Polynesia and Papua New Guinea, Indonesia,
Indian subcontinent, China, Japan, Korea,
Northwest Asia and Siberia, Persian Gulf, Europe, North America,
(South America was discussed in a previous post.
C haplogroup across Southeastern Asia, Australia and Polynesia
To get your bearings in our regional analysis, the following map will come in handy. It shows the frequencies of the different C haplotypes in East and Southeast Asia, PNG, N. Zealand, Polynesia and Australia:
The map clearly shows how hg. C is present in the region at noticeably differing frequencies and with very distinct haplotypes in the each of the geographic locations:
- C3 (M-217). North Asia (Mongolia, Korea, China) and the Americas
- C1 (M-8). Only found in Japan and Ryukyu
- C*. In South China and S.E. Asia, but this is a Paragroup and may conceal yet unknown haplotypes.
- C2 (M-38). From East Indonesia and across Melanesia, Polynesia and New Zealand, but with two distinct haplotypes in the West and the East of this vast region.
- C4 (M-347). Unique to Australia
- C5 (M-356). Not shown in the map, but unique to India, Nepal and Pakistan
As you can see, there is a clearly geographic distribution of haplotypes with little or no overlap.
The following table gives some numerical data for the whole of Asia (including Greater India, not shown in the map above and North America):
This of course mirrors what we have mentioned above: discrete regional distributions and an overlaid C* paragroup at low frequencies (highest in South East Asia) which will have to be resolved into other new haplotypes once their markers have been identified.
Below we will review each region in detail and then try to reach some conclusions.
The Australian Aboriginals - C4
Austrailian natives have their own unique haplogroup, C4, defined by SNP M347; C4 has two haplotypes: C4a (with STR DYS390 deleted) and C4b with M210.
This is the original, ancient haplogroup that arrived in Australia during its peopling wave, which took place some 50 kya and was followed by "considerable isolation after the initial arrival" [4].
The original migrants came from the southern tip of mainland Asia: "Sunda" (encompassing the main Indonesian islands and Malaysia). The "Wallace Line", a strip of sea with many minor islands forming "Wallacea" (Sulawesi, Lombok, Sumbawa, Flores, Sumba, Timor, Halmahera, Buru, etc.) separated it from the other landmass of "Sahul", which was formed by Australia and Near Oceania -New Guinea (NG) and Melanesian Island. The migrants trekked across this now submerged land (sea levels rose after deglaciation c. 9 kya) and rafted across the Wallace Line, reaching Australia.
Interestingly, the other C haplogroups and paragroup C* did not enter Australia (one study detected some C* in Arnhem which may reflect modern arrivals), or if they did, disappeared without a trace.
Despite my hopes that Homo erectus admixed with this Paleolithic migratory wave that peopled Australia and New Guinea (NG), the mainstream science does not support this notion: ""local H. erectus or archaic Homo sapiens populations did not contribute to the modern aboriginal Australian gene pool" [4].
One thing that surprised me is that the time it took the human groups to move across Sahul and Sunda was so long in comparison to what orthodoxy attributes to the peopling of America event:
- Australia. "...the migration from southwestern Asia to Australia would have taken <5,200 years ... This migration speed is in the same order of magnitude as estimated for other prehistoric continental settlements" [4]
- Americas. " the Paleo-Indian spread along the entire longitude of the American double continent might have taken even <2000 yr." [5]
Note that to cross America from Alaska to Cape Horn is about twice the distance compared to the trek from Malaysia to Tasmania yet we must believe that they did it in less than half the time. Did the Paleoindians move four times faster than the Australian Aboriginals' ancestors? Something is not quite right with these numbers. Most likely the American peopling event is far too short, but since orthodox science has to have people in Monte Verde, Chile c. 13 kya and they supposedly entered the New World 15 kya., the absurdly short 2 kya figure appears... Another example of fiddling with the dates to make them fit with preconceptions.
C2: Melanesia, Polynesia and New Guinea (NG)
Melanesia and New Guinea (NG) were settled in the same wave that peopled Australia over 42 kya [6], but later waves of humans reached this area and account for over half the Y chromosome haplogroups in the Region. Nevertheless, haplogroup C is widespread and found at a ㅏ% frequency in Indonesia, indicating its ancient origin [3]. It is found with different haplotypes:
- C* [C-RPS4Y*(xM38, M217, DYS390.1del]. This paragroup is the oldest (50 kya) lineage in Sunda and Sahul [6]. It is very rare in Melanesia and only appears in some coastal NG samples.
It is "notably absent from the NG Highlands and Taiwan and the Philippines" [6], reaching only 3.4% in the latter (Taiwan and Philippines are mentioned because they are apparently the ancestral home of many Melanesians, but seemingly not the C* carriers). [8] - C2.
- C2-M38*(xM208). It is almost absent west of the Wallace Line, its highest frequencies are found in East Indonesia, Moluccas Nusa Tenggara (51 - 44%) [9], coastal NG and
Cook Islands.
It is the most common haplogroup in Northwestern NG (34.6%) and the second most frequent haplogroup in New Guinea (avg. 12.8%). Since its STR diversity his highest in NWNG (Bird's Head region) it very likely originated there (or in Wallacea as the oldest hg. (42 - 61 kya) [6] , expanding east across Melanesia and NG into Polynesia (although rarely observed there); and also West, into Eastern Indonesia. [10][8] - C2b-M208. Found at high frequencies in the West Papua highlands and among Cook Islanders, it is old: mean age 46.2 ky [6] but had a much more recent expansion into
Polynesia (5 - 2.2 kya) [8], where it is found at high frequencies (34%) and also among the New Zealand Maori (77%). [11] [10]
Is a low frequency hg. in Northwest NG (2.5%) and NG (avg. 6.8%). [10]
- C2-M38*(xM208). It is almost absent west of the Wallace Line, its highest frequencies are found in East Indonesia, Moluccas Nusa Tenggara (51 - 44%) [9], coastal NG and
Cook Islands.
A variety (originally named C6) with the P55 marker was reported [6] but is now considered as a Private SNP due to lack of positive testers [2] or a familial group of related males [12] so the C6 haplotype has been dropped and reassigned to European lineages. Three other haplotypes are absent in the region : C5, C3 and the Australian Aboriginal C4 [6].
Rest of Indonesia
- C*. Has a patchy distribution across the Asian region, it is found at high frequencies in East Indonesia (29.2% in Flores and 22.8% in Lembata) and, as mentioned above, is absent in Melanesia and Polynesia. Further north it appears in China (Yao, 20%). [3]
- C2-M38*, is absent in Western Indonesia but grows to an average of 33.5% in Eastern Indonesia (from 11% in Sulawesi to 57% in Sumba); it is found at very low frequencies in Polynesia and Melanesia. [3]
The Indian Subcontinent and the C5 clade
Haplogroup C was detected in India and had been initially classified as C* until Sengupta S. et al., (2006) [14] analysed the paragroup and identified a new haplotype, C5-M356. Almost 85% of the Indian C* individuals were assigned to the new subclade. Which is only found in the Indian subcontinent. [14]
Ancient and autochthonous
C5 is a pan-Indian lineage, absent in the rest of the world yet found at very low frequencies in India (1.4%) where it is widely distributed: It "occurs in all linguistic groups and in both tribes and castes. It also occurs in one Dravidian Brahui in Pakistan" [14]. For this reason, it "is an ancient hg... most plausibly arose in situ within the boundaries of present-day India" [14].
Unsurprisingly, The C3-M217 hg. frequent in Eastern and Central Asia has not been detected in India (it did appear in Pakistan though) [14], neither have C2 or C4.
The data regarding C hg. in India and Pakistan is the following [14] (notice the very low frequencies):
- India
- C*-M216 (RPS4Y) 0.27%
- C5-M356 1.51%
- Pakistan
- C3-M217 6.82%
- C5-M356 0.57%
Tamils of Southern India
The Tamil people inhabit southern India and northern Sri Lanka. A study [15] that sampled tribes, some of which are still foragers, found that haplogroup C was present in 19 out of the 31 groups sampled at relatively high frequency of 4.4% (avg). 90% of them were C5 and the remaining 10% were C*. C hg exhibited a high variance (0.80) suggesting a local origin for its prevailing haplotype.
China
C haplogroup is unevenly spread across China, it is scarce along the eastern coast and more frequent in the North, South and West. The C* paragroup is common in the South and East, and C3 in the North and West.
- Southeast. (Yao, 20% - C*). [3]
- Northeast. Heilongjiang (Manchu, 44.0%) and Hezhe 6.7%, Inner Mongolia (Mongolian, 52.2%; Oroqen, 61.3%), Outer Mongolia (52.8%) [2]
- Northwest. Xinjiang (Hazak, 75.5%) [2]
- Center. Miao 1.7%, Hui 6.7%, Tujia 8.2% (C-RPS4Y*), Han 6% (C-M217), Tujia 18,3% (C-M217)[7] [2]
- South. C*: Mulau 9.1% and Shui 6.9% [2]
Neolithic archaeological remains 6.5 - 2.7 kya from West Liao River valley in Northeast China carried the C3e - P53.1 haplotype. These people are believed to have originated in the northern China steppe, a region where extant populations still carry C3e at a 23.8% frequency. [[17]]
Japan and its C1
C haplogroup is found in Japan at low frequencies, and it has a local haplotype, C1 exclusive to Japan. The values are: [18]
- C1 - M105, the local haplotype, 1.5%
- C3 - M217, originated in the mainland, 2.2%
Korea - C3*
The prevalent C haplogroup in Korea is C. The values differ according to the source: [20] indicated a 12.6% frequency of C-RPS4Y (that is C*, very likely C3*). Another [7] indicated a 16.2% frequency of C3* (xC3c) and yet another [14] 9.6% of C3*.
C3* is also found in neighboring Manchuria at moderate frequencies: higher than in Southeast Asia yet lower than Northeast Asia suggesting an expansion from Mongolia or Siberia into Korea.
Siberia and Northwest Asia
The North Asian haplotypes are (always from the orthodox point of view) relatively recent: C3 as a whole (4.1 to 14.9 ky), and its subclusters are even younger still: C3c (1.6 to 5.9 ky) and C3d (0.5 to 2.0 ky). Notice how wide spread these dates are, which shows the uncertainty originated by the mutation rates used in the calculations. [14]
These are the C lineages found in Northern Asia:
- C3c. (M-48) Prevails among Manchurians, Evens, Kalmyks and Evenks. All of which are Mongolic-Tungusic peoples. [19]
- C3d. Is frequent among Mongol speaking peoples: Mongols, Khamnigans and Buryats.
- C3*. Paragroup with high frequencies (+30%) among Koryaks and Mongols it is also found in North America.
Trivia: history
There is a Genghis Khan "star cluster" (part of paragroup C3*) which is said to have originated 1 kya ago in Mongolia and spread by the Khan's relatives due to their "social status" (a neat sinophylic way of saying that the Mongol hordes raped their way across Eurasia). It now ranges from 35% among Mongols to between 3 and 8% among Buryats, Kazaks, Tuvinians, Shors and Altaians. It is between 0.27 to 2.8 ky old, so it may be related to Genghis Khan and his male relatives. [14][19] Nevertheless, the Kereys tribe in Kazakhstan have the highest frequency (76.5%) of this C3* star cluster and it is unlikely that it is due to Genghis Khan's clan. [19] So maybe these people are the original source of it.
Persian Gulf region
This in Southwestern Asia, the region where haplogroup C first appeared in Asia after leaving Africa. It is found at very low frequencies:
Iran [21]:
- C*. 0.1%. Only appears among the Zoroasterians of Yazd province (2.9%).
- C3. 0.4%, is found in 4 out of 20 ethnic groups, from 0.8% (Bandari) to 2.9% (Zoroasterian).
- C5. 0.5%, appears in 3 groups, From 1.5% (Bandari) to 2.8% (Mazandarani). This is the Indian clade, did it back-migrate into Iran recently or is this a relict?
No apparent pattern, just a patchy distribution: C3 and C5 in the north cicum-Caspian area; C3 and C* in central Iran, C3 and C5 by the Persian Gulf. Perhaps the remnants of the ancient dispersal or are they recent movements of people? [21]
Other Gulf Countries: we also have C* (C-M216) at low frequencies in: Oman 3.3%, Saudi Arabia 1,3% and the UAE 1,2%. [21]
European C6
There is a very rare Southern European haplotype within C hg. The nomenclature for this European C-V20 haplotype has changed: it is now named C6 (originally named C7, but since the NG P-55 became a private marker, C6 was reassigned to Europe). It is characterized by markers V20, V7, V86. V182. V184, V219, V222.
Very few samples were known, a recent study [22] found 1 (one) person carrying C6 out of a sample of 1965 individuals! The paper indicates that "Further studies are needed to establish whether C7 [they use the old notation] chromosomes are the relics of an ancient European gene pool or the signal of a recent geographical spread from Asia.". If the latter, the C6 hg. has yet to be identified in Asia; I am inclined towards an ancient origin in Europe.
Archaic La Braña C6 individual
C6 has been identified in the 7,000 year old remains of a Mesolithic man, discovred at the La Braña site in Spain, supporting its ancient origin. However, with the usual caution of a scientific paper, the authors indicate that: (bold mine):
"... La Braña 1 sample belongs to either haplogroup C or F. When mutations defining those haplogroups were checked, only ancestral alleles were found in the haplogroup F-defining mutations, whereas seven C-defining mutations (M130, M216, P255, P260, V183, V199 and V232) showed only derived alleles. Thus, La Braña 1 most likely belonged to haplogroup C [...] The fact that we found ancestral alleles in mutations defining C1, C2, C3 and C4 (Table S9), together with their actual phylogeographic distribution restricted to Asia, Oceania and the Americas suggests that our individual does not belong to any of these branches. Rather, a new branch within haplogroup C (C6, originally named C7) has recently been identified in several men from Southern Europe, suggesting this could be an ancient European clade. Importantly, mutation V20 showed one read with the derived allele (A), which points to C6 as the most probable sub-clade for La Braña 1 sample. It could also be possible that this G to A mutation is a result of DNA damage. Other less likely haplogroup affiliations are C* and C5 (no read covered SNP M356), both found mainly in present-day India." [23]
North American C3b
Haplogroup C is present in North America at moderate frequencies in a unique haplotype found only in that part of the New World: C3b (P39). The details of the frequencies among the native people: [24]
Tanana (Alaska): 41.7%, Cheyenne: 15.9%, Sioux: 11,4%, Apache: 14.6%, Navajo: 1.3%. It was not detected among any other population across North or Central America.
See my previous post on C3* in South American natives.
Analysis and Discussion
Above I pointed out that Y chromosome hg C is absent in Africa; this means it originated outside of Africa. Conventional mainstream science will therefore place this origin after the OoA (Out of Africa) migration of Modern Homo sapiens some 60 kya.
1. Origin
The accepted theory is that Haplogroup C originated with the split from the hypothetical Haplogroup CF -or CF(xDE). Its marker is SNP P143, which is ancestral to F and C hgs. This split took place somewhere in Southwestern Asia, perhaps on the shores of the Persian Gulf 60 kya.
2. Dispersal across Asia
Men carrying hg. C are believed to have taken an eastern "coastal" route along the coast of the Arabian Sea, reaching the mouth of the Indus River. I see no objections to the possibility that they also advanced inland along the main rivers of this area, but the official dispersal theory sticks to a coastal route (perhaps to tie in the timing of the OoA event and the peopling of Australia, a quick march along the coasts of Asia is required).
2.a. India
From the Indus, C entered the Indian subcontinent, we can suppose that the typically Indian C5 arose later, from those who stayed behind in India, because it is unlikely that C mutated and only those with the C5 marker stayed in India and all the rest, with the non-mutated version kept on moving. Another option is that the mutations arose later from some other region and back- ispersed into India, where it is now prevalent. But this needs us to explain why it became lost in its point of origin.
2.b. Into Austronesia
These migrants pushed on south towards Cape Comorin and Sri Lanka, and then north along the Bay of Bengal and across modern Bangladesh, Myanmar and down into Malaysia, till they reached the edge of the emerged continental shelf in Indonesia: (during the Ice ages sea level was lower so all East Indonesian islands were joined into the Sunda landmass). They boated across the Wallace Line, a stretch of deep seas that blocked access of placental mammals southwards into Australia and New Guinea (NG) -as well as marsupial migrations towards the north. They finally reached Sahul the joined continent of NG and Australia.
The map above shows the current continental area and the emerged continental shelves (in grey), the red arrow shows the migration into Sahul.
It was during these moves that C2 appeared in Wallacea (Western Indonesia and NG) and stayed there while C4 appeared in Australia and also stayed there... why? what kept them from expanding and overlapping in a unified landmass?
The answer: Culture and Topography. Swamps, jungle, valleys and mountain ranges, deserts, rugged shores have kept NG people physically isolated. Tribal societies with their cultural imprint also kept them separated. Current language diversity is a clear indicator of isolation in NG. It is likely that these factors plus population bottlenecks may have kept PNG and Australian natives isolated after the initial single-wave peopling event allowing them to develop their own specific haplotype mutations without any furhter admixture. (Further reading on the peopling of Sahul, McEvoy et al., 2010).
Only much later did another migratory spasm take C2 across the vast Pacific Ocean to people Polynesia and New Zealand but this was a new C2 subclade (C-M208).
2.c. Northwards
We have not considered that these people may have crossed India via the Narmada (by this river, remains of H. erectus were unearthed) and Ganges Rivers or advanced upstream and inland along the Indus, Sutlej, Brahmaputra, Irrawaddy or Saleween rivers, reaching Tibet and Central China or that they may have gone across the continent from Dhaka (Bangladesh) to Hanoi (Vietnam) along the Tropic of Cancer (yellow arrow in map above) and from there advanced inland too (Mekong and Pearl rivers).
No, we have supposed that they took the long tortuous coastal route proposed by orthodoxy, which, in an Ice Age World is shown above (red arrow along Sunda and Blue one northwards towards S.E. Asia). Don't ask me why we must stick to the coast.
I bet that if populations in the interior, in North of Myanmar, Thailand, Yunnan and Laos are sampled, C* and maybe C5 will appear at low frequencies.. maybe this is the homeland of C* from which all others radiated and C5 back-migrated into India.
The following map shows all these possible routes (actually these make more sense than the coastal route for the North Asian populations):
The Green route is the official route. In blue, my suggested change, following the rivers (this explains why it is not so frequent along the East China Sea), a radically different route but also feasible is the Red route, across the Indus, Tian Shan, Altai and Southern Siberia into Manchuria.
I will cite a very interesting paper by Derevianko and Shunkov (2011) [27] which deals with the OoA theory it is worth reading it:
"Early human migration was a slow process, not a relay race. It is hard to conceive why the migrants should have moved directly to the east along the narrow coastal line rather than exploring the banks of the rivers which flow into the ocean and thus moving far to the north, where favorable ecological niches were available." [27]
Having said this, we will assume that, as per Zhong et al., [19] they moved in a "single coastal northward expansion route... in China about 32 to 42 thousand years ago."
2.d. China
They reached Southern China, and the paragroup C* formed here some 36 kya; the migrants continued along the coast (once again we must explain why C* stayed behind and the others moved on), part entered Taiwan (and from there Japan), the others and kept on towards North China leaving barely a trace in the coastal areas of Eastern China (now isn't that strange?).
In North China (Liaoning, Heilongjiang) the C3 haplotype appeared (33 to 20 kya) and dispersed widely: Eastwards, from Manchuria into Korea and Japan. West into Mongolia and South-Central Siberia, Western China, Altai, etc. (20 to 8 kya), and finally as the ices receeded at the end of the last Ice Age, (15 kya) Northwards into Northeast into Eastern Siberia, Beringia and... finally, America.
Another group entered Japan via Ryuku from Taiwan and originated the local C1 haplotype there.
And this is the end of the Orthodox dispersal version.
3. Sub-haplogroups
We have seen above that C hg. has various discrete subhaplogroups (C1, C2, C3, C4, C5 and C6 plus a paragroup C*) each with a specific geographical distribution. Common sense indicates that they "have undergone long-time isolation" [2]. But little is said about how they originated without spreading into other regions.
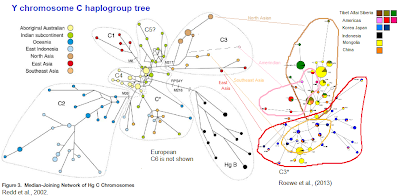
Below I adapted Fig. 2, from Redd A., et al., (2002) [26]; at that time the C5 haplotype from India was not known and the C4 among Australians was not shown in the tree, so I added the C4 dotted line around the yellow Aboriginal dots; I also wrote C5? around the green Indian dots within C* paragroup (top part). C3 in this figure does not include Native Americans and surely contains a lot of paragroup C3*. So I included on the right hand side, the C3* data for Asia and America from Roewer et al., (2013) [19], correlating the regions to the color code of Redd et al. (I added the South American natives in pink), but I maintained the individual data dots with their original colour (key is on upper right corner).
Also see C3* phylogenetic tree in my previous post.
The tree is quite revealing as it shows a central core of C* from which the other branches appear. Actually, a closer look at the tree reveals that this central C* as per Redd et al., is Indian, and it is linked to the probable root in the form of the branch joining it with haplogroup B (on the left).
The most central branches of the tree are the Australian C4 (left-center) tightly gathered in the middle, and Indian C5 (center and top-center) with the other branches located further apart:
- C2. (bottom left). Is diverse as can be judged from its spread branches, and it arises from C4
- C1. (upper left). Is quite diverse and arises from C*
- C3. (upper right). Is diverse and is also born from C*
- C*. (middle and bottom center). Is diverse. The main branch on the bottom is mainly South East Asian, but there are also some Indian in it. It is very spread and closely linked to the root of C2 and C4, which suggests that this South Asian C* and the Austronesian lineages C2 and C4 are the very old lines of the C hg. peopling wave.
Comments:
The bottom clusters are basically a widely divergent C2 sprouting from the Australian C4, and C* in S.E. Asia
The top clusters are C1, C5 and C3 all sprouting from the archaich core.
Redd's original root from Hg. B, anchors in Indian C* (green dots). I have no way of telling apart the C5 and the C* of (See data above: Indian origin), my guess is that the upper cluster is genuine C5 while the green dots in the central part of the tree and the bottom are actually C*.
The core from which C1, C5, C3, C* and C4 sprout from are green Indian subcontinent dots... surely C* from India. Only C2 is clearly rooted in one line of C4
1. Origins
C2. Arose in Wallacea from an ancestral lineage of C (which must be the original C M-130 variety) carried by the group that would move into Australia and Melanesia. Part of them moved on into Australia forming C4, the other proto-C people remained in Wallacea and formed derived clades: paragroup C-M38* and haplogroup C-M208, which remained in this territory located East of the Wallace Line and much later moved into Polynesia. Topography, tribal structure and bottlenecks kept C2 and C4 apart.
C4. See above, the "ancestral C stock" from Wallacea entered Australia and remained there in isolation.
Of course the ancestra C line is ancestral to current C* in S. E. Asia, and the C4, C5, C1 and C3 lines. C* is still found in India and other parts of S.E. Asia, it is represented by the green dots in the central part of the image. These will prove to be a new haplotype (H7?) when their marker is found. This is the ancestral lineage, from which all others sprout, it is Indian.
This may help explain some of the questions we posed during the haplotype analysis:
C5 originated from this ancestral C in India, the ancestral C people are the original peopling wave, they moved north and became C3 and C1, they moved south and became C4 in Australia and C* in S.E. Asia, a yet to be discriminated haplogroup (H8?) It
The European C6 is not placed in the tree but it is surely a group that split early in Asia and marched West into Europe.
Clearly further analysis is necessary to breakdown C* into newer sub-clades.
C* in Indonesia has a very high STR variance: this means that it has had plenty of time to mutate; it is very old. In Indonesia it is more frequent in the East, because a "continual eastward migration of the initial settlers (i.e., settlements were not permanently established in western Indonesia) or later waves of (partial) replacement." overlaid it. [3] This corroborates is antiquity.
As we can see in the Table of C haplotype distribution, above, haplotype C3 is concentrated in Northern Asia and has a decreasing cline towards the south (absent in Australia, NG and Polynesia) and west, it is found in North America at relatively high frequencies (C3b - P39, unique to the New World) and in South America in a patchy C3* distribution (see my previous post on C3* in South America.
A sinophile paper [19] proposes that this cline is due to its Chinese origin some 42 to 32 kya, and a coastal north route of expansion, it adds that the highest STR diversity for C3 is found in Southeast Asia. This, in my opinion corrobrates its origin from the ancestral C group, but not in China; it appeared in a region that is central to the other Haplotypes:
The region currently occupied by Myanmar, North Thailand, East India and Yunnan in China.
From there it irradiated and became the current regional haplotypes.
Some Crazy ideas
Having given the official story and facts, allow me to let my imagination fly and suggest an alternative scenario based on these same facts.
The area where C hg. is found with a highest diversity in Asia is precisely the area where Homo erectus lived for over 1.5 million years: Southern and Eastern Asia (from China and Korea in the North, to India and Indonesia in the south).
Haplogroup C's coastal route is precisely the one supposedly taken by our distant H. erectus ancestor.
The OoA theory with H. sapiens originating in Africa and peopling the world, totally replacing previous extant populations (if they existed), is so widely accepted that it dealt a death blow to the multiregional theory (H. sapiens evolved in different regions from a common archaich ancestor). But recent genetic discoveries of admixture with Neanderthals, Denisovans and mysterious "X" hominins as well as some remains in China with a mosaic of archaic and modern features have put fresh wind in the sails of the Multiregional theory.
For instance, Derenko and Shunkov (2011) question the OoA theory and support the Multiregional hypothesis in which an "independent formation of anatomically modern humans occurred", this took place in three regions with four subspecies all of which merged into modern humans:
- East and South East Asia, with Homo sapiens orientalensis
- Rest of Eurasia, with Homo sapiens neanderthalensis and Homo sapiens altaiensis (Denisovans)
- Africa, with Homo sapiens africanensis
Among other things, they also use the evidence of a distinct post-Middle Paleolithic South East Asian lithic industry that was different to that of Europe and Asia, with an "autochthonous development of the Upper Paleolithic,"[27]
They go a step further and suggest a Homo erectus evolution into modern humans in Asia:
"...population of anatomically modern humans descended from Homo erectus locally. .... progressive biological traits are due to parallel evolution. Both in East Asia and in Africa, anatomically modern humans apparently originated from the same ancestral species – Homo erectus sensu lato. ...The totality of evidence speaks in favor of a progressive in situ evolution of Homo erectus in East Asia over a span of more than one million years. This does not preclude the immigration of small populations from adjacent regions, small-scale gene flow, or admixture." [27]
If this is the case, then the C haplogroup found among South East Asians is in fact the one carried by H. erectus from Africa into Asia and later mutated in situ in Asia.
But this would contradict the accepted notion that our Y chromosomes are exclusively human, and that our ancestors (and their Y chromosomes) split from those of H. erectus long ago. Their genes disappeared with them and therefore do not appear in us.
But a study has already suggested an ancient origin for human Y chromosome: (Mendez et al, 2013) it reports the discovery of a novel Haplogroup named A00, which gave a very old age: "338 thousand years ago (kya) (95% confidence interval = 237-581 kya). Remarkably, this exceeds current estimates of the mtDNA TMRCA, as well as those of the age of the oldest anatomically modern human fossils..." [16]. In other words this "human" Y chromosome haplogroup is older than humans!
Which of course has been criticised by OoA proponents as using incorrect mutation rates that pushed the dates too far into the past.
I wonder, (read my previous posts criticising the mutation rate calculations) if actually the mutation rates for this (and All other) haplogroups is not underestimated by a factor of three or four, which would mean that the dates could be 711 - 2,381 ky, more than enough to accomodate H. erectus in the picture.
This means that Y chromosomes mutate far slower than currently accepted. And that when we look at the current distribution of NRY hgs. we are seeing the ancient migrations of pre-sapiens men across the globe.
Current C hg. distribution reflects the migration of Homo erectus out of Africa 1.8 Mya. A band of a few hundreds of people walking into Asia with the CF haplogroup, splitting in the Persian Gulf by acquiring the M130 marker, and thus forming C haplogroup. They moved across South Asia keeping their C hg. identity during their long trek (note that since mutation rates are slower than accepted, no mutations arose during this period).
Finally reaching the Homeland from which it differentiated into its current haplogroups, in the North of S. E. Asia. From there they spread out. Some moved into NG, Australia mutating (slowly) into C2 and C4. Others went back into India and mutated to C5. The core in S.E. Asia evolved into C* while others went north forming C1 and C3.
A group did not take the Eastern route and went West into Europe forming C6 there. Maybe it will be sequenced someday from the bones at Sima de los Huesos...
Erectus entered America long ago, and the patchy C3* distribution in South America is what remains of a once widespread coverage of H. erectus in the New World, and not a recent transpacific junk with Jomons from Japan shipwrecked on the shores of Ecuador.
Of course this could be proved or rejected if DNA could be sampled and sequenced from H. erectus remains (maybe impossible due to DNA decay). Maybe in the future it could be done, who knows?
Sources
[1] Chuan-Chao Wang and Hui Li, (2013). Inferring human history in East Asia from Y chromosomes. Investigative Genetics 2013, 4:11 doi:10.1186/2041-2223-4-11
[2] Hua Zhong et al., (2010). Global distribution of Y-chromosome haplogroup C reveals the prehistoric migration routes of African exodus and early settlement in East Asia. Journal of Human Genetics doi: 10.1038/jhg.2010.40
[3] Tatiana M. Karafet et al., (2010). Major East–West Division Underlies Y Chromosome Stratification across Indonesia. Mol Biol Evol (2010) 27 (8): 1833-1844. doi: 10.1093/molbev/msq063 First published online: March 5, 2010
[4] Georgi Hudjashov et al., (2007). Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis. vol. 104 no. 21, 8726–8730, doi: 10.1073/pnas.0702928104
[5] Martin Bodner, Ugo A. Perego et. al., (2012). Rapid coastal spread of First Americans: Novel insights from South America's Southern Cone mitochondrial genomes. Genome Res. May 2012; 22(5): 811–820. doi: 10.1101/gr.131722.111
[6] Laura Scheinfeldt, (2006). Unexpected NRY Chromosome Variation in Northern Island Melanesia. Mol. Biol. Evol. 23(8):1628–1641. 2006. doi:10.1093/molbev/msl028
[7] Yali Xue, et al., (2006). Male demography in East Asia: a north-south contrast in human population expansion times Genetics 172:4 (April 2006): pages 2431-2439.
[8] Stephen Oppenheimer, (2006). The 'Austronesian' story and farming-language dispersals: Caveats on timing and Independence in Proxy Lines of Evidence from the Indo-European Model, from "Uncovering Southeast Asia's Past: Selected Papers from the 10th International Conference of the European Association of Southeast Asian Archaeologists : the British Museum, London, 14th-17th September 2004" European Association of Southeast Asian Archaeologists. NUS Press, Jan 1, 2006
[9] Kayser M, Underhill P, et al., (2003). Reduced Y-chromosome, but not mitochondrial DNA, diversity in human populations from West New Guinea. Am. J Hum Genet 72:281–302
[10] Stefano Mona et al., (2007). Patterns of Y-Chromosome Diversity Intersect with the Trans-New Guinea Hypothesis. Mol Biol Evol (2007) 24 (11): 2546-2555. doi: 10.1093/molbev/msm187 First published online: September 10, 2007
[11] Underhill PA, Cavalli-Sforza LL., et al., (2001). The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62
[12] Y-DNA Haplogroup C and its Subclades - 2014. International Society of Genetic Genealogy
[13] Asian Ancestry based on Studies of Y-DNA Variation: Part 1 Early origins – roots from Africa and emergence in East Asia. Genebase Tutorials. http://www.genebase.com/learning/article/21
[14] Sanghamitra Sengupta., et al., (2006). Polarity and Temporality of High-Resolution Y-Chromosome Distributions in India Identify Both Indigenous and Exogenous Expansions and Reveal Minor Genetic Influence of Central Asian Pastoralists. Am J Hum Genet. Feb 2006; 78(2): 202–221 Dec 16, 2005. doi: 10.1086/499411
[15] Ganesh Prasad Arun Kumar et al., (2012) Population Differentiation of Southern Indian Male Lineages Correlates with Agricultural Expansions Predating the Caste System. PLoS ONE 2012. doi:10.1371/journal.pone.0050269
[16] Mendez et al., (2013). An African American paternal lineage adds an extremely ancient root to the human Y chromosome phylogenetic tree. Am J Hum Genet. 2013 Apr 4;92(4):637.
[17] Yinqiu Cui et al., (2013). Y Chromosome analysis of prehistoric human populations in the West Liao River Valley, Northeast China. BMC Evolutionary Biology 2013, 13:216
[18] Nonaka, I., Minaguchi, K. and Takezaki, N., (2007). Y-chromosomal Binary Haplogroups in the Japanese Population and their Relationship to 16 Y-STR Polymorphisms. Annals of Human Genetics, 71: 480–495. doi: 10.1111/j.1469-1809.2006.00343.x
[19] Roewer L., et al., (2013). Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans. PLoS Genet 9(4): e1003460. doi:10.1371/journal.pgen.1003460
[20] Soon Hee Kim, Myun Soo Han, Wook Kim, and Won Kim, (2010). Y chromosome homogeneity in the Korean population. International Journal of Legal Medicine 124:6 (November 2010): pages 653-657.
[21] Grugni, V. et al., (2012). Ancient Migratory Events in the Middle East: New Clues from the Y-Chromosome Variation of Modern Iranians. PLoS ONE 7(7): e41252. doi:10.1371/journal.pone.0041252
[22] Scozzari R, Massaia A, D’Atanasio E, Myres NM, Perego UA, et al. (2012) Molecular Dissection of the Basal Clades in the Human Y Chromosome Phylogenetic Tree. PLoS ONE 7(11): e49170. doi:10.1371/journal.pone.0049170
[23] Iñ,igo Olade, et al., (2014). Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European, Nature (2014) doi:10.1038/nature12960
[24] Stephen Zegura, Tatiana M. Karafet et al., (2004). High-Resolution SNPs and Microsatellite Haplotypes Point to a Single, Recent Entry of Native American Y Chromosomes into the Americas. Molecular Biology and Evolution, vol 21(1), pp 164-75
[25] Moss, S. J. & Wilson, E. J. 1999. Biogeographic implications of the Tertiary palaeogeographic evolution of Sulawesi and Borneo. In Hall, R. & Holloway, J. D. (eds) Biogeography and Geological Evolution of SE Asia. Backhuys Publishers, Leiden, 133-155.
[26] Redd, Alan J., et al., (2002). Gene Flow from the Indian Subcontinent to Australia: Evidence from the Y Chromosome. Current Biology, Vol. 12, Issue 8, 16 April 2002, Pages 673–677. doi: 10.1016/S0960-9822(02)00789-3
[27] A.P. Derevianko and M.V. Shunkov, (2011) Anthropogenesis and colonization of Eurasia by Archaic Populatoins. Formation of anatomically Modern Human. From
Proceedings of the International Symposium “Characteristic Features of the Middle to Upper Paleolithic Transition in Eurasia: Development of Culture and Evolution of Homo Genus”
(July 4–10, 2011, Denisova Cave, Altai). Edited by A.P. Derevianko, M.V. Shunkov. pp 50 - 74.
Patagonian Monsters - Cryptozoology, Myths & legends in Patagonia Copyright 2009-2014 by Austin Whittall ©





On Y-DNA hg C as well as the new clade MP, see the recent paper buy Magoon et al. Some comments on Dienekes's are informative, too: http://dienekes.blogspot.com/2013/11/a-priori-y-chromosome-phylogeny-from.html. I think we're moving toward a phylogeny whereby C3 is opposed to all the other Cs. Take a read. It's remarkable that all Amerindians, including the paragroup C3* carriers don't fall out of the C3 cluster and it's the one that may be opposed to all others.
ReplyDeleteI keep getting fascinated by the fact that Amerindians belong to only two clades - C3 and Q which bookend the whole non-African phylogeny. No transitional clades have been found in America.
Ust-Ishim's Y-DNA haplogroup has been established but not publicly revealed.
German,
ReplyDeleteYes indeed, C3* is a very odd branch within C. It spans Eurasia and the Americas, and is, in my opinion very very ancient.
Thanks for the link and the tip on Y hg.
The fact that C and Q are the only ones in America is definitively not due to them being at the end of the last OoA branch. No. These are ancient haplogroups.
I am also wondering if some of the R in the Americas came early, that is before the European discovery in 1492.
How can it be told apart? Every time R hg. is found among natives they are believed to be admixed with recent European migrants!!
Ust-Ishim will be of particular interest because of its age. If there were modern H. sapiens in Siberia 45 kya, which admixed with Neanderthals some 200 to 400 generations earlier... this means that they moved quickly OoA or they left Africa earlier than currently accepted. Will he be P, Q, R? (is it a he or a she?), what mtDNA?
ReplyDeleteUst-Ishim results haven't been published yet and I can't disclose them. But what has already become known via a leaked tweet from a conference is that Ust-Ishim mtDNA belongs to hg R. Notably, all the most ancient mtDNA samples (Tianyuan, Mal'ta, Kostenki) are hg R, too. Hg R has the widest distribution among modern populations (which under a certain population model would make it the oldest macrohaplogroup), although it's usually portrayed as the youngest of the macrohaplogroups. No African-specific haplogroups have been found in ancient remains outside of Africa and without this kind of evidence out of Africa lacks proof.
ReplyDeleteWhat's intriguing about Ust-Ishim is that Amerindians follow Ust-Ishim in having the longest chunks of Neandertal DNA (see chart at http://dienekes.blogspot.com/2014/04/svante-paabo-talk-at-nih.html). Ust-Ishim is 45,000 years old. Amerindians clearly didn't admix with Neandertals 20-15,000 years ago (as 696 generations would imply), hence the rate of decay of those chunks is not constant and Amerindians must be a "genetically conservative" population and not young at all.
If Ust-Ishim is R then it is really intersting. As I mentioned further up, I have this nagging feeling that part of the R found in America is ancient, far older than Vikings or the sixteenth century voyages of discovery, but... how can that be proved?
DeleteUst-Ishim man have an mtDNA Hg R* and Y DNA Hg K2-M526.
DeleteHello~ Mr.Whittall. this is the latest information on C HaplogroupTree
ReplyDeleteC - M216
C1 - F3393
C1a - CTS11043
C1a1 - M8 Japan
C1a2 - V20 Europe
C1b - F1370
C1b1 - M359 Hindu-Arabic Coast
C1b2 - M38 Lesser Sunda Islands + Pacific islands
C1b3(?) - (Tentative, Do not confirmed) M347 Australia
C2 - M217
C2a - M93 Japan
C2b - L1373
C2b1 - P39 Anglo America
C2b2 - M48 Kamchatka Peninsula
C2b3 - F1396 Turco-Mongol
C2c - P53.1 Tungus
C2d - P62 Mongolia
C2e - Z1338 East Asia (C2e accounts for more than 90% of all C haplogroup. but they were not Nomadic peoples)
Thank you for the updated nomenclature. I see that the Japanese M8 and the European V20 are in the C1a branch. That is very intriguing.
DeleteThen the C1b which encompasses the southern route all the way to Polynesia (Persian Gulf, India, Indonesia, PNG, maybe Australia...)
C2 which is the Asian-American branch.
Excuse me, AW. Can you tell me more information about Y Chromosome Haplogroup C2e-Z1338 and it's distribution?
DeleteThis comment has been removed by the author.
ReplyDeleteSo, An Eurasian Adam M168 give rise a Men with Haplogroup DE* YAP+ and Haplogroup CF*-P145. Interesting!
ReplyDeleteHaplogroup C1a and C1b have now been found in ancient DNA samples dating back over 30ka (one in Belgium, one in Russia, one in Czech Republic).
ReplyDeleteAlso, there are over 250 defining mutations for C*, which means that from its divergence with Haplogroup CF, there is a period of over 20ka in which it is completely unattested. Plenty of time to originate about anywhere before its two branches appear. It seems like a lot more research is needed.