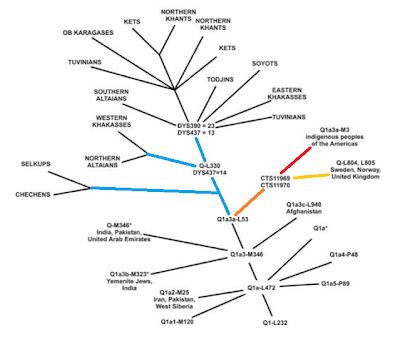
I am following up on some (there were quite a few) of the subjects that appeared while writing my most recent posts on the human Y chromosome.
Among them, are my very serious doubts regarding the markers used in classifying Y-chromosome haplogroups, the ages of the different sub-clades or haplotypes within them, and the different dates for common ancestors based on the estimated mutation rates for the human Y chromosome. As expressed in my previous posts, the methods, assumptions and convoluted statistical calculations seem far fetched and very uncertain. The fact that some confidence intervals are too large in my opinion invalidate the logic behind them, but then, I am a mere engineer not a geneticist.
The Y chromosomes of the Neanderthal
It is in this context that I read an interesting paper (Musaddeque Ahmed and Ping Lian, 2013) [1], from which I quote the following regarding Neanderthal Y chromosomes ... (bold font is mine):
"...the Y-chromosome of Neanderthals or paternal inheritance has yet to be examined. Comparisons of the Y chromosome sequence of Neanderthals with currently established Y-haplogroups for modern humans should provide some insights into the admixture hypothesis.
With respect to the recent finding of admixture of Neanderthals with non-African populations, the Neanderthal Y chromosome should not match the Y haplogroups A or B, as these haplogroups are the oldest of the clades and almost restricted to Africans and their descendants. Since haplogroup E is found in Africa, the Middle East, Southern Europe, and Asia, the Neanderthal Y chromosome may match this haplogroup, but it should not match the haplogroups E1b1a*, E2b1, or B2a1a, as they are specifically treated as Bantu expansion markers, while Neanderthals interbred only with non-Africans." [1]
I was very surprised by this paragraph, as I expected something similar to what we see in mtDNA, where the ancestral population of both humans and Neanderthals, split into two groups: one that would later evolve into modern humans, and another that would evolve into Neanderthals. Each would carry, in the beginning, the "original" archaic Y chromosome of the ancestral population, which, as each lineage accumulated mutations due to chance and positive selection would begin to grow in different directions, forming two distinct branches, which then in turn would continue branching as more mutations appeared.
So, the oldest human Y-chromosome haplogroups "A" and "B" found exclusively in Africa, were born from the branch leading to modern humans (the "root" of mankind), while, on a separate branch, (Neanderthals), the Y chromosome haplogroups of Neanderthals appeared.
However, towards the end of this post we will see that this reasoning may not be correct.
Nevertheless, mainstream science considers it to be the correct interpretation. According to Krause et al. (2007), [2] since the oldest common ancestor with human Y chromosomes dates back to 90 kya, and as Neanderthals and humans split long before that date, they "expect[ed] Neandertal Y chromosomes to fall outside the variation of modern human Y chromosomes unless there was male gene flow from modern humans into Neandertals"; in other words: Neanderthals would have their own peculiar Y chromosome haplogroups unless H. sapiens men mated with Neanderthal women and passed on their Y chromosomes to their human-neanderthal mixed male sons. But then these would not be pure Neanderthals but admixed ones, with 50% human genome in them and their Neander mom's mtDNA.
The following image shows what I expressed above, two branches: red for Neanderthals and black for us, humans, both splitting from our common ancestor:
Actually, we have not yet found Neanderthal Y chromosomes among modern human genomes and this is indeed curious since we have admixed with them. It could be due to several reasons:
(a) They are present in such low frequencies that they have not yet been sampled. Imagine a 0.001% frequency, that is, only 1 in 100,000 men would carry it (35,000 in the whole world,
a very small number indeed).
(b) They are very similar to modern haplogroups (as suggested by Musaddeque and Ping, 2013. [1]) and we see them but have not yet realized that they are Neanderthal.... (More on this
below, because it seems to be a very interesting possibility).
While studying the Neanderthal genome, Krause et al., (2007) [2] found that two of their Sidrón Neanderthal specimens (Sidrón 1253 and Sidrón 1351c) were males. They then tested them at the five positions that mark the main nodes of Human branches (i.e. the splitting points of our Y chromosome haplogroups) in both Eurasia and Africa. They noticed that "all 15 Y chromosomal products for the five assayed positions show the ancestral allele" [2]. In other words, the ancestral or chimpanzee-like state was found. They did not find any specifically Neanderthal-like state. This is odd, considering that 7 million years separate chimps from Neanders, why would they both share the markers in their Y chromosomes and humans, a mere 300 ky away don't. It may be an error in their sequencing.
Admitting the paucity in their sampling and that these Y-chromosomes may appear in extremely low frequencies among modern human populations, the authors conclude:"These [Neanderthal] Y chromosome results must derive, then, either from Y chromosomes that fall outside the variation of modern humans or from the very rare African lineages not covered by the assay..." [2], in other words, the Neanderthal Y chromosomes may be similar to those of modern humans that were not screened for.
I must point however, that according to Figure S1, the "markers" chosen by Krause et al., screened only certain branches of the Y chromosome tree: (i) and (ii) the split that leads to all haplogroups downstream from the A clade. (B, C,.... R2). (iii), The one leading to B. (iv) The one leading to P, (and therefore Q), R1, R2. And (iv) the one leading to A2.
So the Neanderthal Y chromosomes could belong to Haplogroups A1 and A3, which were not screened for. Or, an option that is evident, but they did not consider it at all: Neanderthal Y chromosomes are identical to those of Modern Humans and cannot be told apart from them.
Actually, isn't it surprising that not one Neanderthal Y (or X with its mtDNA) chromosome survived until our days? We have a considerable amount of autosomal genetic content introgressed from our relatives, the Neanderthals, but their mithocondria and their sex determining chromosomes did not survive. Why?
An extremely old Y chromosome lineage
Mendez et al., (2013) [3] , discovered a Y chromosome haplogroup (named A00) which shares its most recent common ancestor with the rest of mankind 338 kya, of course (as usual!) the confidence interval is 237 to 581 kya. So this is a very old and wide time frame which goes back well beyond the oldest known fossils of anatomically modern human (AMH) beings.
This haplogroup was found in the genes of an African American and later identified in the chromosomes of Mbo individuals from Cameroon, at very low frequencies: 0.19% [CI = 0.11%–0.35%].
They would predate the oldest modern human fossils, which are only 195 ky old. So the Mbo lineage should have split from humanity at least 40 ky before our species even appeared.
The paper also looked at the rest of our Y chromosome lineages, working up the other branches of the tree, finding that the African A0 hg diverged from the other branches 202 kya (CI = 125 - 382 kya), which is much older than previous estimates of 142 kya.
This in turn moves the split between African and non-African branches (hg C to T) backwards in time, making them older: 63 kya vs. the previous 39 kya date.
Orthodoxy strikes back
Of course, such an ancient root for modern humans is inconsistent with mainstream genetic beliefs so a rebuttal appeared (Eran Elhaik et al., 2014)[4], pointing out that it: "contradicts all previous estimates in the literature and is over 100,000 years older than the earliest fossils of anatomically modern humans.".
E. Elhaik et al believe that Mendez et al.'s paper overestimated mutation rates resulting in an artificially older age. Their analyisis "indicates that the A00 lineage was derived from all the other lineages 208300 (95% CI= 163900 - 260200) years ago." [4] Which as can be seen, falls suitably close to the oldest dated AMH fossils.
New May 2018: Recent findings have pushed back both the earliest H. sapiens remains' datings in Africa and in Asia. This means that an older origin for AMH is indeed possible.
But let's get back to the very ancient A00 and the "unorthodox" point of view:
What are the implications of this finding? There are several possible scenarios:
1. Archaic admixture. These Mbo people inherited their A00 haplogroup from an achaic human population. These ancient African humans carried the A00 lineage in them, mated with AMH (within the last 195 ky) and passed on their Y chromosomes to their male offspring, their A00 passed from father to son in these AMH, which survived until now (Mbo's are their youngest descent). In the meantime, the archaic group became extinct.
Mendez et al. support this view: "Interestingly, the Mbo live less than 800 km away from a Nigerian site known as Iwo Eleru, where human skeletal remains with both archaic and modern features were found and dated to ~13 kya." [3]
2. Deeper time line. Maybe the depth of modern humans into the past is greater than the current (spotty) fossil record shows (roughly 200 kya).
The Confidence intervals given by Mendez et al., span from 237,000 to 581,000 years ago, this is well into the dark past of mankind, a time frame which encompasses many hominids in Eurasia and Africa: H. erectus (from 1.8 Mya until roughly 300 kya); Denisovans (until 30 kya), Homo heidelbergensis (600 to 400 kya), Neanderthals (600 to 25 kya) and H. rhodesiensis (300 to 125 kya).
Could this Y chromosome lineage belong to one of these hominids? Before answering let's just consider one additional piece of information.
Chuan-Chao Wang et al., [5] noticed that Y-chromosome STR haplotypes from different haplogroups converge; they looked for "possible haplotype similarities among haplogroups [and found] similar haplotypes between haplogroups B and I2, C1 and E1b1b1, C2 and E1b1a1, H1 and J, L and O3a2c1, O1a and N, O3a1c and O3a2b, and M1 and O3a2 ..." [5] .
In other words identical Short Tandem Repeat (STR) markers crop up among the different Y chromosome haplogroups indicating that the same mutations arise time and time again, unlike the markers of haplogroups which are much more sporadic and unique.
Making sense of this muddle
Above, I asked why didn't Neanderthal Y chromosomes survive among their descent (albeit admixed with modern humans)? Probably there were too few of them and too many of us, and their Y chromosomes got watered down, in successive admixtures, diluted until their frequencies are so low that they have not yet been detected.
Another option is that "Haldane's rule'' kicked in; the rule declares that if hybrids of one sex only are sterile, the afflicted sex is much more likely to be the male (XY) than the female (XX). Of course, why would hominds so close to each other bear sterile offspring? It is not like horses and donkeys, this is humans and Neanderthal, and we have their genes in us, so we did interbreed and the offspring survived (but Haldane's rule may mean that the girls survived but the boys were sterile).
A paper (Sankararaman et al., 2014) [6] supports the "Haldane's rule" notion: "...interbreeding of Neanderthals and modern humans introduced alleles onto the modern human genetic background that were not tolerated, which probably resulted in part from their contributing to male hybrid sterility".
The Neanderthal-deficient regions in modern humans are found in genes that "are specifically expressed in the testes, and in the female sex X chromosome. "This suggests that some Neanderthal-modern human hybrids had reduced fertility and in some cases were sterile. An unexpected finding is that regions with reduced Neanderthal ancestry are enriched in genes, implying selection to remove genetic material derived from Neanderthals. Genes that are more highly expressed in testes than in any other tissue are especially reduced in Neanderthal ancestry, and there is an approximately fivefold reduction of Neanderthal ancestry on the X chromosome." [6]
But, this does not mean a total extinction of their sex chromosomes; not all of their offspring were sterile after all, we still carry their genes mixed with ours, why shouldn't their Y chromosomes survive too?
In my opinion, we actuall carry their Y chromosomes in us (in the men of course), but long before we admixed with them in Eurasia some 60 kya. The mutation rates that are used to date our Y lineages are wrong, allow me to explain why:
We look at populations (say Amerindians) and jot down their haplogroup markers, and we assume that they mutated when they reached America and then, we guess the date they entered America (say 15 kya). With this we calibrate our clock. We again look at the humans closest to Africa and jot down their haplogroups markers, and once again guess the date these people's ancestors left Africa (say 70 kya), and again calibrate our clock. We take another look at the oldest fossils of AMH in Africa (195 kya) and jot down the most divergent African haplogroups' markers, we recalibrate our clock again. But, as you can see, there are many assumptions in all of this (the dates and, above all, the assumption that these haplogroups are specifically human and mutated recently < 200 ky!).
But, What if they are not specific to us, but archaic? What if we carry slowly mutating Y chromosomes. The mutations found in certain haplogroups are valid, but they reflect ancient migrations. Maybe even the Out Of Africa (OOA) migration of H. erectus or, within the time range given by Mendez et al., the Y chromosomes of H. heidelbergensis or Neanderthals?
This would explain why there are no Neanderthal specific branches to be found (the red ones in the image above). We all have the same tree our and their lineages coincide.
So what the date should we consider for the makers at the "non-African" CT groups (the OOA split)? Not the date modern humans left Africa. Instead it may be the date Neanderthals left Africa.
The split in Altai, with a Western route for our "R" hg and East for the "Q" hg may indicate divergence among the Neanderthals that lived there while AMH began to move out of Africa.
The archaic humans of China and even Lake Mungo people in Australia may be the branches of the South East Asian and Austronesian Y chromosome haplogroups, instead of modern humans that reached those regions much later.
The Q hg in America may not reflect a recent peoping of America at all, but an ancient one by Neanderthals.
The pan African E hg, may reflect the recent dispersal of AMH in the continent as well as in the Middle East, Southern Europe, and Asia after their OOA movement.
The dispersal of Q hg across the Arctic regions of America and Eurasia or the presence of the very old C hg in Asia and America may reflect the ancient migrations of primitive humans and not the recent (<20 kya) dispersal of modern humans.
See This post for an update. May 2018 and this Sept. 2020 update
Sources
[1] Musaddeque Ahmed and Ping Lian, (2013). Study of Modern Human Evolution via
Comparative Analysis with the Neanderthal Genome. Genomics Inform. Dec 2013; 11(4): 230–238. Published online Dec 31, 2013. doi: 10.5808/GI.2013.11.4.230
[2] Johannes Krause, et al., (2007). The Derived FOXP2 Variant of Modern
Humans Was Shared with Neandertals. Current Biology 17, 1–5, November 6, 2007 ª2007 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2007.10.008
[3] Fernando L. Mendez et. al., (2013). An African American Paternal Lineage Adds an Extremely Ancient Root to the Human Y Chromosome Phylogenetic Tree. The American Journal of Human
Genetics 92, 454–459, March 7, 2013. http://dx.doi.org/10.1016/j.ajhg.2013.02.002.
[4] Eran Elhaik, T. Tatarinova, A. Klyosov and D. Graur, (2014). The 'extremely ancient' chromosome that isn't: a forensic bioinformatic investigation
of Albert Perry's X-degenerate portion of the Y chromosome. European Journal of Human Genetics (2014) 1-6. doi:19.1038/ejhg.2013.303
[5] Chuan-Chao Wang et al., (2013). Convergence of Y chromosome STR haplotypes from different SNP haplogroups
compromises accuracy of haplogroup prediction. pre-print Arxiv server on 21 October 2013.
[6] Sriram Sankararaman et al., (2014). The genomic landscape of
Neanderthal ancestry in present-day humans. Nature 507, 354–357 (20 March 2014) doi:10.1038/nature12961
Patagonian Monsters - Cryptozoology, Myths & legends in Patagonia Copyright 2009-2018 by Austin Whittall ©